Bioactive compounds often bind to several target proteins, thereby exhibiting polypharmacology. Experimentally determining these interactions is however laborious, and structure-based virtual screening of bioactive compounds could expedite drug discovery by prioritizing hits for experimental validation.
The Pharmaceutical Bioinformatics group at the University of Freiburg has therefore developed Zauberkugel, a workflow for pharmacophore-based protein target prediction of a bioactive ligand with the tool Align-it and the recently reported pharmacophore dataset ePharmaLib. The Zauberkugel workflow requires only two inputs; the ligand structure file (SMI format) and the ePharmaLib dataset (PHAR format). The latter can be retrieved from Zenodo.
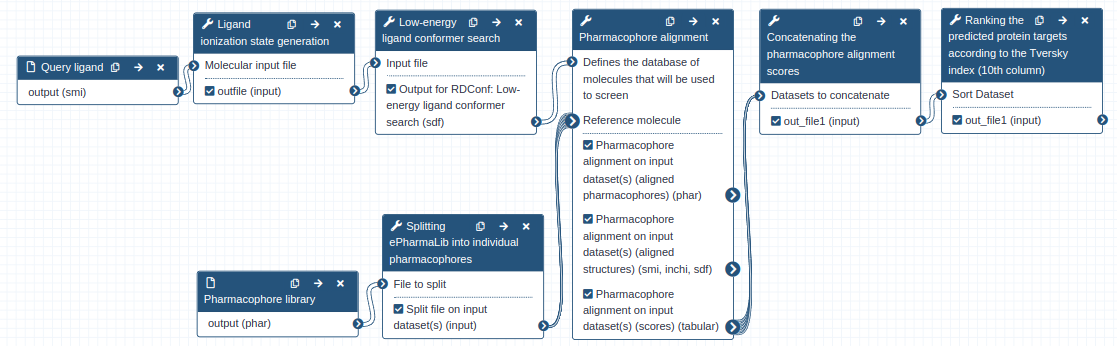
To learn more on how to use the Zauberkugel workflow, please refer to the Galaxy training material.
Credit
ePharmaLib: A Versatile Library of e-Pharmacophores to Address Small-Molecule (Poly-)Pharmacology
Aurélien F. A. Moumbock, Jianyu Li, Hoai T. T. Tran, Rahel Hinkelmann, Evelyn Lamy, Henning J. Jessen, and Stefan Günther.
Journal of Chemical Information and Modeling 2021, 61(7), 3659-3666.
https://doi.org/10.1021/acs.jcim.1c00135
