Methods to read out naturally occurring or experimentally introduced nucleic acid modifications are emerging as powerful tools to study dynamic cellular processes. The recovery, quantification and interpretation of such events in high-throughput sequencing datasets demands specialized bioinformatics approaches.
One of such methods is SLAMseq, a sequencing technology for time-resolved measurement of newly synthesized and existing RNA in cultured cells. Originally developed by the lab of Stefan Ameres, the lab of Johannes Zuber extended the approach with pharmacological and chemical-genetic perturbations in order to identify direct transcriptional targets of any gene or pathway (Muhar et al, Science 2018).
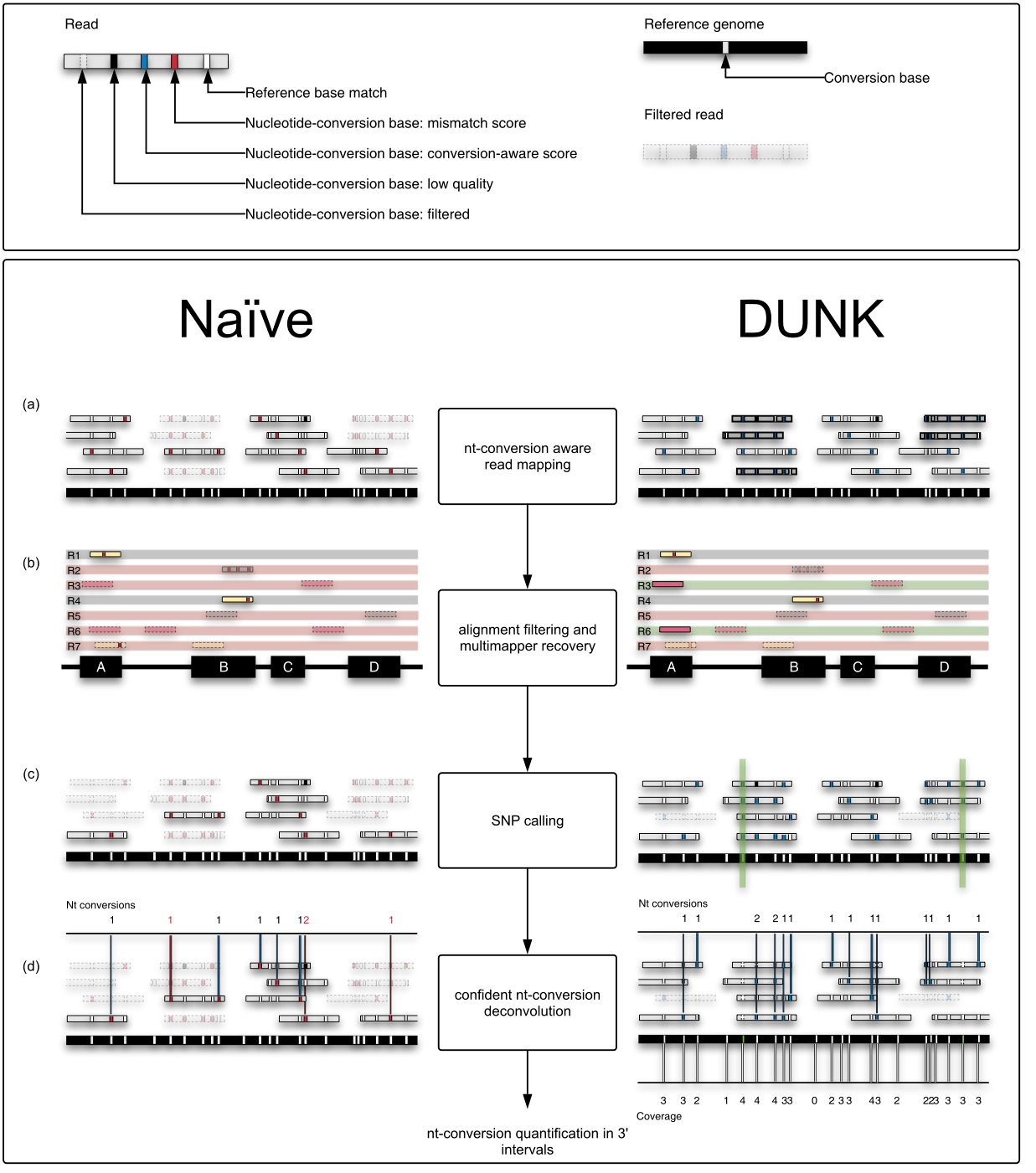
Processing and interpreting this data required novel analysis methods, so we developed Slamdunk which we recently published in BMC Bioinformatics and is generally applicable to any nucleotide-conversion containing dataset.
Workflows
Two workflows have been developed for Slamdunk and are available
-
Slamdunk: This workflow quickly produces, from raw SLAMseq data, robust and reproducible nucleotide-conversion quantifications in a set of given genomic intervals
-
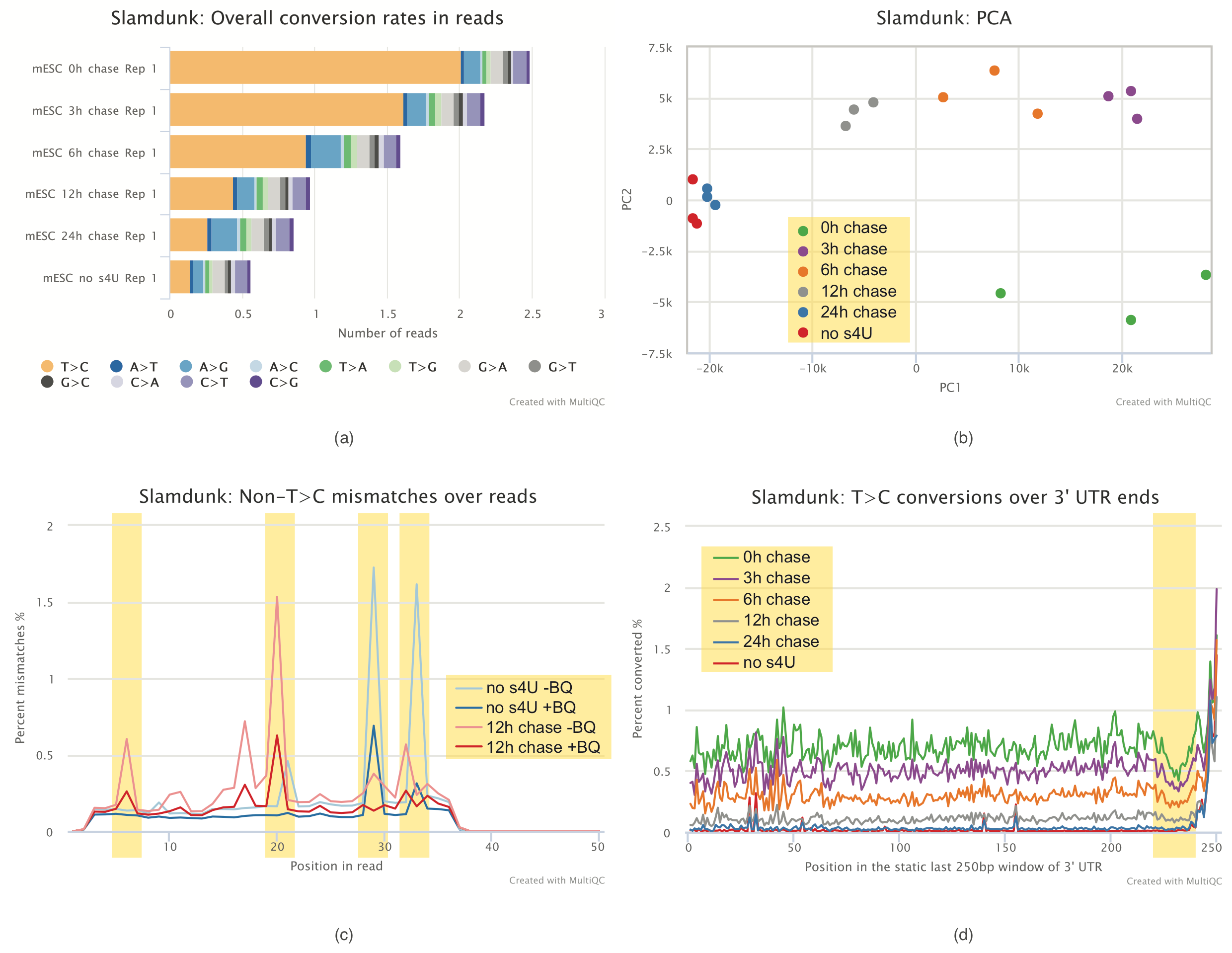
Alleyoop: This analysis will run several QC tools on the produced SLAMseq datasets from Slamdunk and present in a ready-to-go MultiQC report as shown below.